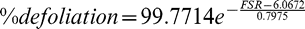Introduction
Tomato spotted wilt, caused by Tomato spotted wilt virus (genus Tospovirus; family Bunyaviridae) (TSWV) and transmitted by thrips (Thysanoptera: Thripidae), is a serious problem to southeastern U.S. peanut production. Tomato spotted wilt occurs every year with fluctuations in severity within years and locations (Culbreath et al., 2003). Losses to TSWV in peanut increased steadily from the late 1980s, with annual losses as high as $40 million in Georgia alone (Bertrand, 1998). TSWV is transmitted by several species of thrips (Ullman et al., 1997), but the control of thrips has not shown much promise for management of spotted wilt in peanut. The management of spotted wilt is dependent upon the use of cultivars with moderate levels of field resistance (Culbreath et al., 1996), and cultural practices which reduce the incidence and severity of spotted wilt (Brown et al., 1996; Culbreath et al., 1999). New cultivars such as AP-3 (Gorbet, 2007), Georgia-02C (Branch, 2003) , and Tifguard (Holbrook et al., 2008a) have greater levels of field resistance than Georgia Green, the moderately resistant cultivar used as a standard for genotype comparisons (Culbreath et al. 2008), but still can be severely affected. Higher levels of field resistance have been reported in the cultivar Georganic (Culbreath et al., 1997; Culbreath et al., 1999; Holbrook and Culbreath, 2008) and breeding line F NC94022-1-2-1-1-b3-B (Culbreath et al., 2005; Li et al., 2009), henceforth referred to as NC 94022.
Two of the most prevalent and severe foliar diseases of peanut worldwide are early leaf spot, caused by the fungus Cercospora arachidicola S. Hori and late leaf spot, caused by the fungus Cercosporidium personatum (Berk. & M. A. Curtis) Deighton (Smith and Littrell, 1980). In the U. S., most of the peanuts are grown in the southern states where environmental conditions are often favorable for leaf spot epidemics. One or both of the leaf spot diseases occur in all peanut producing states, and multiple applications of fungicides are necessary for control of the diseases.
The development and use of resistant cultivars is one of the most desirable ways to manage both leaf spot diseases and TSWV (Culbreath et al., 2003). Currently, peanut breeding programs focus on extensive field screening to look for sources of resistance and making selections of resistant lines once crosses have been made and breeding lines are available. Field selection for resistance to leaf spot pathogens and TSWV requires considerable space and a relatively large number of plants. Development of molecular markers for assisting in selection for resistance to TSWV and the leaf spot pathogens could increase the efficiency of breeding programs (Li et al., 2010; Guo et al., 2012). Marker assisted selection has been used in development of nematode resistant peanut cultivars COAN (Simpson and Starr, 2001) and NemaTAM (Simpson et al., 2003), but markers are not currently available for resistance to either of the leaf spot pathogens or TSWV. The objective of this study was to compare field susceptibility of diverse peanut genotypes, including cultivars, germplasm accessions, and advanced breeding lines to TSWV and leaf spot pathogens in order to relate to genotypic characterization of these lines in an effort to develop markers for resistance to TSWV and leaf spot pathogens (Li et al., 2010) through genetic mapping (Qin et al., 2012).
Materials and Methods
Twenty-two genotypes were evaluated in this study, including commercial cultivars, advanced breeding lines, and germplasm accessions (Table 1). Two separate field trials were conducted at the University of Georgia, Coastal Plain Experiment Station Belflower Farm, Tifton, GA in 2007 and 2008. Soil type is Tifton loamy sand (Fine-loamy, siliceous, thermic Plinthic Kandiudult). Peanut was in an annual rotation sequence following corn (Zea mays L.). In each year, one experiment was planted in April to maximize potential for development of spotted wilt epidemics (Culbreath et al., 2003), and one was planted in May to reduce potential for spotted wilt epidemics and increase the likelihood of heavy leaf spot epidemics.
The field trials were conducted using randomized complete block designs with 4 replications. Experiment plots were 6.0 m long, separated by 2.4-m alleys. Peanut seed was planted to 91-cm-spaced twin-row plots. Planting dates were 23 April and 23 May in 2007, and 18 April and 20 May in 2008. Seeding rate was 3.3 seed/m of row. Sparse seeding rate was used to maximize pressure of Tomato spotted wilt (Culbreath et al., 2003) and to allow Tomato spotted wilt severity evaluation on individual plants. Only April planted trials were used for the spotted wilt evaluations, and both April and May planting trials were used for leaf spot evaluations. Fungicide applications of chlorothalonil (Bravo WeatherStik, Syngenta Crop Protection, Greensboro, N.C.) (1.26 kg a.i./ha) were made at ca. 30 days after planting with subsequent applications every 14 days until ca. 80 days after planting.
Tomato spotted wilt severity was evaluated for each plant on 25 July, 2007 and 21 July, 2008. Severity of Tomato spotted wilt was assessed using a 0 to 5 severity scale adapted from Baldessari (2008) based on visual determination of presence of symptoms and estimation of the degree of stunting (reduction in plant height, width, or both) for symptomatic plants (Table 2). Genotype comparisons were made for total incidence of Tomato spotted wilt (severity ratings of 1 or greater), as well as incidence of plants with severity ratings in classifications (2 or greater, 3 or greater, 4 or greater and 5).
Leaf spot severity was evaluated on 25 September 2007, and 26 September 2008 for each plot using the Florida 1 to 10 scale (Chiteka et al., 1988) where 1 = no leaf spot; 2 = very few lesions on the leaves and none on upper canopy; 3 = very few lesions on upper canopy; 4 = some lesions with more on upper canopy, and 5% defoliation; 5 = noticeable lesions on upper canopy with 20% defoliation; 6 = numerous lesions on upper canopy with 50% significant defoliation; 7 = numerous lesions on upper canopy with 75% defoliation; 8 = Upper canopy covered with lesions with 90% defoliation; 9 = very few leaves covered with lesions remain and some plants completely defoliated; 10 = plants dead (Chiteka et al., 1988). Final percent defoliation (% defoliation) was estimated from the Florida scale rating taken just before peanut plants were inverted (Chiteka et al., 1988).
Final leaf spot ratings were converted to estimates of % defoliation according to the function:
This equation was obtained by non-linear regression of defoliation level on Florida scale rating (FSR) values as reported by Chiteka et al. (1988).
Data were analyzed using Proc Mixed with ddfm = satterth option that performs a general Satterthwaite approximation for the denominator degrees of freedom and produces an accurate F-approximation (SAS v.8.3, SAS Institute, Cary, NC), unless otherwise stated. Incidence of Tomato spotted wilt and % defoliation by leaf spot estimated from Florida scale ratings were used as response variables. Genotype was considered a fixed variable, and replication and year were treated as random effects. Main effects and interactions as well as specific treatment effects were considered significant if P ≤ 0.05. Fisher's least significant difference (LSD) was computed using standard error and t values of adjusted degrees of freedom from ddfm = satterth option. If the interaction effect between year and genotype was not significantly different (P > 0.05), the data were pooled and presented averaged across years; if the interaction was significant (P ≤ 0.05); the data were analyzed separately and are presented by year.
Genotypes were classified as susceptible, moderately susceptible, moderately resistant, and resistant, based on TSWV incidence and leaf spot intensity compared to that in Georgia Green. As an interesting note for 2007 and 2008 growing seasons, early leaf spot was the predominate disease in the field and we rated the total defoliation including both early and late leaf spots.
Results and Discussion
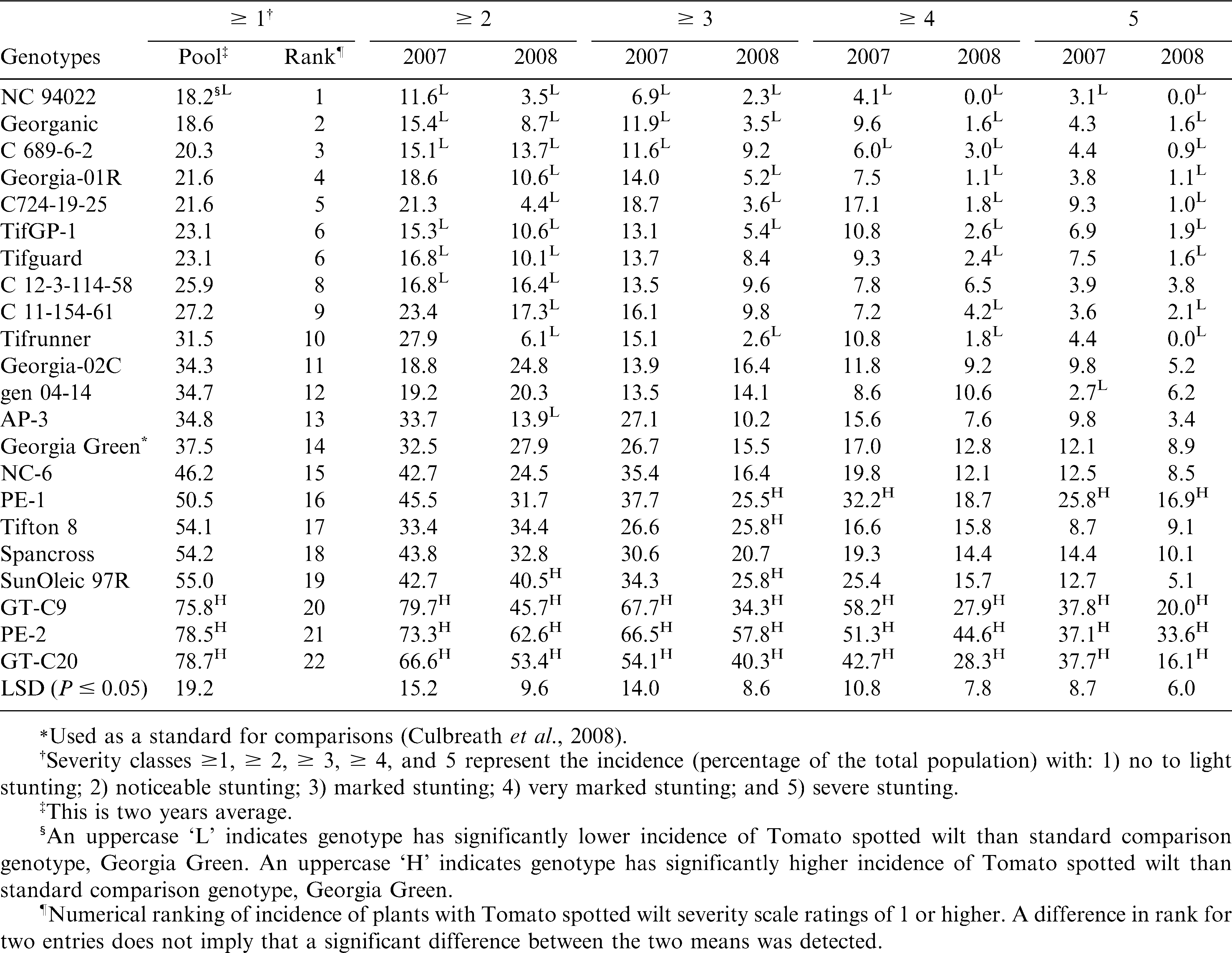
There was no significant interaction between year and genotype for the total incidence of Tomato spotted wilt (severity ≥ 1), but there were significant interactions for the incidence of severity categories, ≥ 2, ≥ 3, ≥ 4 and 5 (data not shown). Therefore, data were pooled across years for the analysis of incidence of severity ≥ 1, but incidence of plants in all other severity ratings categories are presented separately for each year.
For the incidence of severity ≥ 1, NC 94022 had the lowest numerical ranking for total incidence of spotted wilt, but did not differ from that of thirteen genotypes (Table 3). Only NC 94022 had total incidence of spotted wilt (severity rating ≥ 1) lower than that of Georgia Green (Table 3). For other categories of incidence of severity, NC 94022 had lower incidence of Tomato spotted wilt than Georgia Green in both years (Table 3). Several additional genotypes in addition to NC 94022 had incidence of Tomato spotted wilt that ranked lower than Georgia Green in all severity classes (Table 3). Among them, only U.S. breeding line C689-6-2 showed a lower incidence of Tomato spotted wilt than Georgia Green in all severity classes for at least one year (Table 3). Others including U.S. released cultivars Georganic, Georgia-01R, Tifguard, Tifrunner, AP-3 and breeding lines C724-19-25, TifGP-1, C 12-3-114-58, C11-154-61 showed significantly lower incidences of Tomato spotted wilt than Georgia Green in one or more severity classes in each year (Table 3).
GT-C20, which originated from China, had the highest rating in total incidence (Table 3). There were no significant differences among GT-C20, GT-C9 and PE-2, and they were all greater than Georgia Green in all severity classes in both years (Table 3). Incidence of Tomato spotted wilt in SunOleic 97R, Tifton 8, and PE-2 typically ranked higher than that of Georgia Green and was significantly higher in one or more severity classes in each year (Table 3).
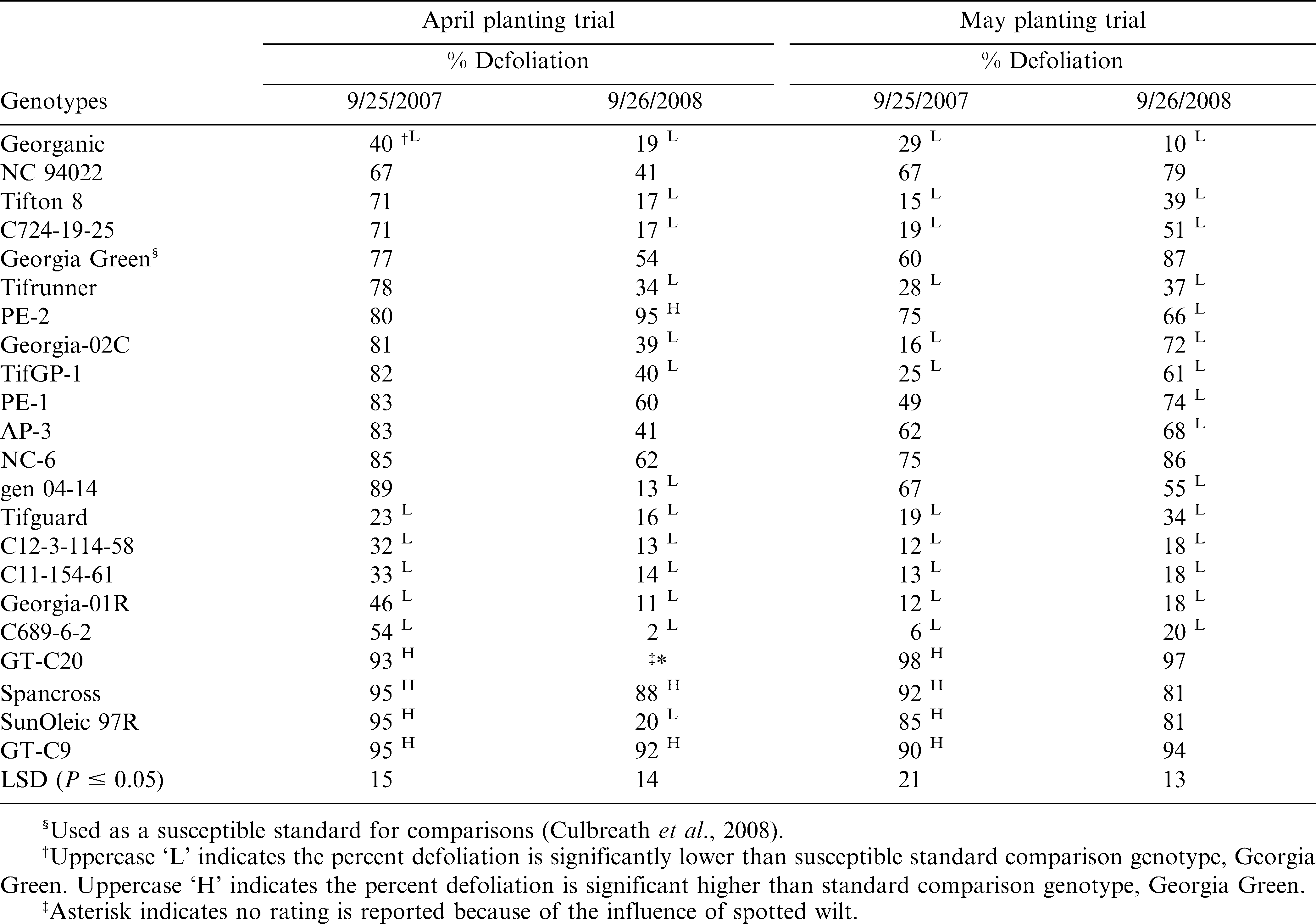
Leaf spot epidemics were severe in both trials in both years and caused noticeable defoliation in all genotypes evaluated (Table 4). In 2008, leaf spot evaluations were not made in GT-C20 in the early planted trial because of severe stunting and mortality from Tomato spotted wilt by the time of leaf spot evaluation. A significant year by genotype interaction for percent defoliation for both early and late planted trials was observed; therefore, means of defoliation are presented by trial and year.
Defoliation for the cultivars Georganic, Georgia-01R and Tifguard, and breeding lines C12-3-114-58, C11-154-61 and C689-6-2 was significantly lower than for Georgia Green in both early and late planted trials for both 2007 and 2008; whereas, defoliation for the cultivars Tifrunner and Georgia-02C and breeding lines C724-19-25 and TifGP-1 was significantly lower than Georgia Green in late planted trials for both years and for the early planted trial in 2008 (Table 4). U.S. breeding line gen 04-14 had significantly less defoliation than Georgia Green for both trials in 2008, but not in 2007. Several additional genotypes including AP-3 and PE-1 had lower defoliation than Georgia Green for the late planted trial in 2008 (Table 4).
Defoliation for GT-C20 and SunOleic 97R was greater than for Georgia Green in both trials in 2007, and defoliation for Spancross and GT-C9 was greater than for Georgia Green for both trials in 2007 and the early planted trial in 2008. Except for GT-C20 and SunOleic 97R, there were no other genotypes that showed higher defoliation than Georgia Green for the late planted trial in 2008. In general, the late planted peanuts had lower levels of defoliation by leaf spot than early planted peanuts. However, the leaf spot evaluations for both tests were made on the same day. Therefore, the different levels of leaf spot in these two tests likely were due to longer epidemic duration in the early-planted trails than the late planted trials.
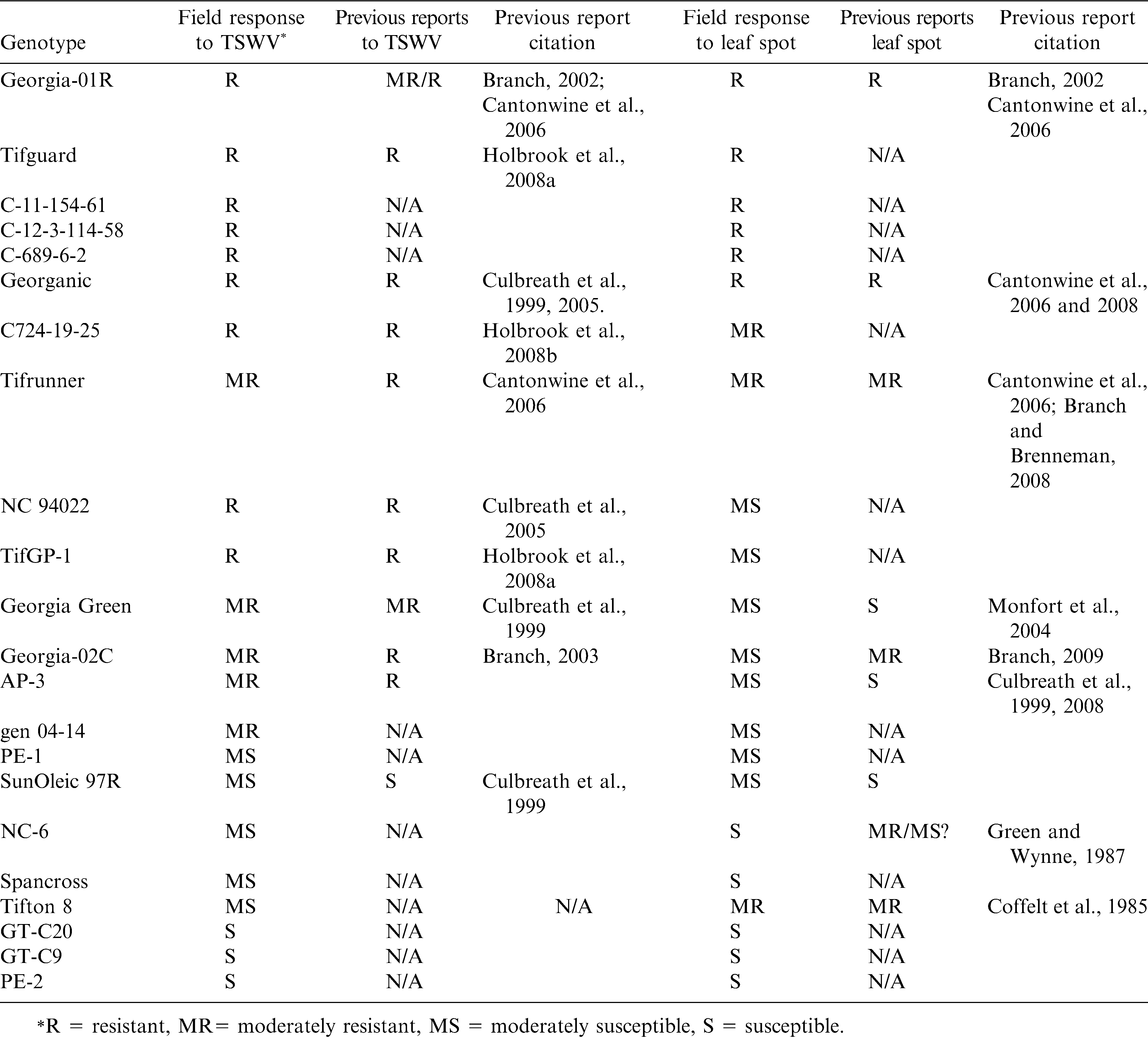
A summary of the disease responses of the genotypes in the current study, compared to previous reports of resistance to each pathogen, is listed in Table 5. The results from this study generally agreed with previous reports and comparisons of current results to previous reports indicated that the field resistances to TSWV in NC94022 (Culbreath et al., 2005), Georganic (Culbreath et al., 1999; Culbreath et al., 2005), Tifguard (Holbrook et al., 2008b), C724-19-25 (Holbrook et al., 2008c), Tifrunner (Holbrook and Culbreath, 2007), and Georgia-01R (Culbreath et al., 2008) were better than that in Georgia Green (Table 5). SunOleic 97R was reported as susceptible to TSWV (Culbreath et al., 1999; Culbreath et al., 2005). In the current study there were genotypes from China that showed higher severity of Tomato spotted wilt disease compared to that of SunOleic 97R as susceptible to TSWV. Three other U.S. cultivars, NC-6, Spancross, and Tifton8, and one Chinese breeding line PE-1 showed similar incidence of Tomato spotted wilt in the fields as genotypes with moderate susceptibility (Table 5). Field reaction to Tomato spotted wilt had not been reported previously for most of the breeding lines including, C689-6-2, TifGP-1, C12-3-114-58, and C11-154-61, which showed promising levels of field resistance to TSWV. Genotypes PE-2, GT-C9, and GT-C20 were highly susceptible to TSWV (Table 5).
In general, there were three cultivars (Georgia-01R, Tifguard, and Georganic) and three breeding lines (C 11-154-6, C 12-3-114-58 and C 689-6-2) classified as resistant to both TSWV and leaf spots. Whereas, some Chinese lines, GT-C20, GT-C9 and PE-2 were highly susceptible to both TSWV and leaf spots. PE-1 was moderately susceptible to both TSWV and leaf spots (Table 5). There has been no report of Tomato spotted wilt in peanut in China; therefore, resistance to TSWV would not have been selected by Chinese breeders. For those genotypes from China, leaf spot evaluations in the current study were hindered by high incidence and severe symptoms of Tomato spotted wilt. However, susceptibility to leaf spots was obvious.
The results in this study were also in agreement with previous reports of field resistance to leaf spots in Georgia-01R (Culbreath et al., 2008), Georganic (Culbreath et al., 1999; Culbreath et al., 2005) and Tifrunner (Holbrook and Culbreath, 2007) (Table 5). Tifguard and its sister line C725-19-25 showed resistance to moderate resistance to leaf spots in this study. A preliminary report from another study indicated Tifguard also had a moderate level of resistance to late leaf spot (Culbreath et al., 2008). Although field resistance to TSWV in NC 94022 has been reported (Culbreath et al., 2005), this study evaluated the reaction of NC94022 to both leaf spot pathogens and TSWV. NC 94022 proved to be moderately susceptible to early leaf spot in this study but has been observed to be resistant to late leaf spot which was the predominant leaf spot in 2010 (Culbreath and Guo, personal communication). Among the tested breeding lines, three lines, C11-154-61, C12-3-114-58 and C689-6-2, were identified as resistant genotypes showing lower percent defoliation due to leaf spot, whereas C724-19-25 and TifGP-1 were moderately resistant.
Summary
The results in this study indicated a wide range of field reactions to TSWV and leaf spots among the tested lines. This information should be useful for formulating disease management strategies by using different peanut cultivars or breeding lines. In addition, these trials included fourteen of a panel of sixteen genotypes for which genetic diversity was characterized using simple sequence repeat (SSR) markers (Li et al., 2010) and four of these genotypes, NC94022, SunOleic 97R , Tifrunner, and GT-C20, have been used to develop two recombinant inbred line (RIL) populations (Qin et al., 2012). Based on the results of these field trials, the parents, NC 94022 and SunOleic 97R differ markedly in their field resistance to TSWV, while the other pair of parents Tifrunner and GT-C20 differs greatly in field resistance to both TSWV and leaf spots. Information from the combination of the field and genetic characterization of these genotypes could be useful in development of mapping populations and construction of genetic linkage map for quantitative trait loci (QTLs) studies (Qin et al., 2012).
Acknowledgements
We thank Billy Wilson and Jake Fountain for technical assistance in the field. This research was partially supported by USDA Specific Cooperative Agreement 58-6602-6-121 with the University of Georgia, and the graduate student assistantship was partially supported by funds provided by Georgia Peanut Commodity Commission and the National Peanut Board. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Literature Cited
Baldessari J.J 2008 Genetics of Tomato spotted wilt virus resistance in peanut (Arachis hypogaea L.) University of Florida , Gainesville, FL 113 pp.
Bertrand P.F 1998 . 1997 Georgia plant disease loss estimates University of Georgia Coop. Ext. Serv. Pub. Path 98-007 , Athens, GA .
Branch W.D 2002 Registration of ‘Georgia-01R’ peanut Crop Sci. 42 : 1750 – 1751 .
Branch W.D 2003 Registration of ‘Georgia-02C’ peanut Crop Sci. 43 : 1883 – 1884 .
Branch W.D and Brenneman T.B 2008 Field evaluation for the combination of white mod and Tomato spotted wilt disease resistance among peanut genotypes Crop Prot. 28 : 595 – 598 .
Brown S.L Todd J.W and Culbreath A.K 1996 Effect of selected cultural practices to Tomato spotted wilt virus and populations of thrips vectors in peanuts Acta. Hortic. 431 : 491 – 498 .
Cantonwine E.G Culbreath A.K Holbrook C.C and Gorbet D.W 2008 Disease progress of early leaf spot and components of resistance to Cercospora arachidicola and Cercosporidium personatum in runner-type peanut cultivars Peanut Sci. 35 : 1 – 10 .
Cantonwine E.G Culbreath A.K Stevenson K.L Kemerait R.C Brenneman T.B Smith N.B and Mullinix B.G 2006 Integrated disease management of leaf spot and spotted wilt of peanut Plant Dis. 90 : 493 – 500 .
Chiteka Z.A Gorbet D.W Shokes F.M Kucharek T.A and Knauft D.A 1988 Components of resistance to late leaf spot in peanut. I. Levels and variability – implications for selection Peanut Sci. 15 : 25 – 30 .
Coffelt T.A Hammons R.O Branch W.D Mozingo R.W Phipps P.M Smith J.C Lynch R.E Kvien C. S Ketring D.L Porter D.M and Mixon A.C 1985 Registration of Tifton-8 peanut germplasm Crop Sci. 25 : 203 – 203 .
Culbreath A.K Gorbet D.W Martinez-Ochoa N Holbrook C.C Todd J.W Isleib T.G and Tillman B.L 2005 High levels of field resistance to Tomato spotted wilt virus in peanut breeding lines derived from hypogaea and hirsuta botanical varieties Peanut Sci. 32 : 20 – 24 .
Culbreath A.K Tillman B.L Gorbet D.W Holbrook C.C and Nischwitz C 2008 Response of new field resistant peanut cultivars to twin row pattern or in-furrow applications of phorate insecticide for management of spotted wilt Plant Dis. 92 : 1307 – 1312 .
Culbreath A.K Todd J.W and Brown S.L 2003 Epidemiology and management of Tomato spotted wilt in peanut Annu. Rev. Phytopathol. 41 : 53 – 75 .
Culbreath A.K Todd J.W Gorbet D.W Branch W.D Sprenkel R.K Shokes F.M and Demski J.W 1996 Disease progress of Tomato spotted wilt virus in selected peanut cultivars and advanced breeding lines Plant Dis. 80 : 70 – 73 .
Culbreath A.K Todd J.W Gorbet D.W Brown S.L Baldwin J.A Pappu H.R Holbrook C.C and Shokes F.M 1999 Response of early, medium, and late maturing peanut breeding lines to field epidemics of Tomato spotted wilt Peanut Sci. 26 : 100 – 106 .
Culbreath A.K Todd J.W Gorbet D.W Shokes F.M and Pappu, H.R. H.R 1997 Field response of new peanut cultivar UF 91108 to Tomato spotted wilt virus Plant Dis. 81 : 1410 – 1415 .
Gorbet D.W 2007 Registration of ‘AP-3’ peanut J. Plant Registr. 1 : 126 – 127 .
Green C.C and Wynne J.C 1987 Genetic-variability and heritability for resistance to early leafspot in 4 crosses of virginia-type peanut Crop Sci. 27 : 18 – 21 .
Guo B.Z Chen C.Y Chu Y Holbrook C.C Ozias-Akins P and Stalker H.T 2012 Advances in genetics and genomics for sustainable peanut production In Benkeblia N (ed.) Sustainable Agriculture and New Biotechnologies CRC Press , Boca Raton, FL . pp. 341 – 368 .
Holbrook C.C and Culbreath A.K 2007 Registration of ‘Tifrunner’ peanut J. Plant Registr. 1 : 124 .
Holbrook C.C and Culbreath A.K 2008 Registration of ‘Georganic’ peanut J. Plant Registr. 2 : 17 .
Holbrook C.C Timper P and Culbreath A.K 2008a Registration of peanut germplasm line TifGP-1 with resistance to the root-knot nematode and Tomato spotted wilt virus J. Plant Registr. 2 : 57 .
Holbrook C.C Timper P Culbreath A.K and Kvien C.K 2008b Registration of ‘Tifguard’ peanut J. Plant Registr. 2 : 92 – 94 .
Holbrook C.C Timper P Dong W.B Kvien C.K and Culbreath A.K 2008c Development of near-isogenic peanut lines with and without resistance to the peanut root-knot nematode Crop Sci. 48 : 194 – 198 .
Li Y Culbreath A.K Guo B.Z Knapp S.J and Holbrook C.C 2009 Tomato spotted wilt and early leaf spot reactions in peanut genotypes from the U.S. and China Phytopathology 99 : S73 .
Li Y Chen C.Y Knapp S.J Culbreath A.K Holbrook C.C and Guo B.Z 2010 Characterization of simple sequence repeat (SSR) markers and genetic relationships within cultivated peanut (Arachis hypogaea L.) Peanut Sci. 38 : 1 – 10 .
Monfort W.S Culbreath A.K Stevenson K.L Brenneman T.B Gorbet D.W and Phatak S.C 2004 Effects of reduced tillage, resistant cultivars, and reduced fungicide inputs on progress of early leaf spot of peanut (Arachis hypogaea) Plant Dis. 88 : 858 – 864 .
Qin H Feng S Chen C Guo Y Knapp S Culbreath A He G Wang M.L Zhang X Holbrook C.C Ozias-Akins P and Guo B.Z 2012 An Integrated genetic linkage map of cultivated peanut (Arachis hypogaea L.) constructed from two RIL populations Theoretic and Applied Genetics 124 : 653 – 664 .
Simpson C.E and Starr J.L 2001 Registration of ‘COAN’ peanut Crop Sci. 41 : 918 – 918 .
Simpson C.E Starr J.L Church G.T Burow M.D and Paterson A.H 2003 Registration of ‘NemaTAM’ peanut Crop Sci. 43 : 1561 .
Smith D.H and Littrell R.H 1980 Management of peanut foliar diseases Plant Dis. 64 : 356 – 361 .
Ullman D.E Sherwood J.L and German T.L 1997 Thrips as vectors of plant pathogens In: Thrips as Crop Pests , Lewis T (ed.) CAB International , Wallingford, UK . pp. 539 – 564 .
Notes
- University of Georgia, Department of Plant Pathology, Athens, GA 30602
- USDA-Agricultural Research Service, National Peanut Research Laboratory, Dawson, GA 39842
- University of Georgia, Department of Crop and Soil Science, Athens, GA 30602
- USDA-Agricultural Research Service, Crop Genetics and Breeding Research Unit, Tifton, GA 31793
- USDA-Agricultural Research Service, Crop Protection and Management Research Unit, Tifton, GA 31793 *Corresponding author (e-mail: Baozhu.Guo@ars.usda.gov)
Author Affiliations