Introduction
Cultivated allotetraploid (2n = 4x = 40) peanut (Arachis hypogaea L.) is an important oil seed crop worldwide. Peanuts are important both due to the oil produced in the seeds and also the favorable seed composition for promoting positive human health. Chemical and epidemiological studies have consistently shown that peanuts are nutritionally beneficial because they contain lipids, folate, tocopherol, and protein (Dean et al., 2009). Unsaturated fatty acyl residues and their effect on human health have received a lot of attention during the past decade (Vassiliou et al., 2009). For example, oleic acid has been shown to be associated with reducing systolic blood pressure (Teres et al., 2008), reducing triacylglycerol (Pelkman et al., 2004), guarding low density lipoproteins (LDL) from oxidative modification (Parthasarathy et al., 1990), helping maintain good cholesterol levels known as high density lipoproteins (HDL), reducing blood glucose levels in type II diabetes (Vassiliou et al., 2009), and slowing down atherosclerosis (Yu et al., 2008). Further, low fat diets rich in monounsaturated fats are more effective for improving serum lipid and apolipoprotein levels than a low fat diet devoid of monounsaturated fats (O'Byrne et al., 1997).
Fatty acids found in plants are the principal components of membrane phospholipids and triacylglycerol (TAG) storage (Zhang et al., 2009). Generally, peanuts are composed of 45–51% oil in the seed, of which, approximately 80% is composed of the two major fatty acids oleic (C18∶1) and linoleic (C18∶2) [Lopez et al., 2000]. The range of these predominant fatty acids in peanut germplasm can vary, but generally span 36 to 80% for oleic acid and 2 to 43% for linoleic acid (Norden et al., 1987). The fatty acid composition in oil seed crops are, however, a major determinant of their overall quality (Moore and Knauft, 1989; Jung et al., 2000b). The fatty acids influence seed quality due to the likelihood of oxidation of the oil which can break down over time and ultimately produce noxious odors or off flavors in stored food products. Consequently, the shelf life of peanut products is limited. Generally, saturated fatty acids are less prone to oxidation during processing and storage than unsaturated fatty acids (Moore and Knauft, 1989; Lopez et al., 2001) because the double bonds in polyunsaturated fat break down to produce acids, aldehydes, ketones, and hydrocarbons (Moore and Knauft, 1989). Hence, peanut seeds that have a high amount of oleic acid, a monounsaturated omega-9 fatty acid, and a low amount of linoleic acid, a polyunsaturated omega-6 fatty acid, are generally preferred by manufacturers and consumers.
In 1987, the first reported high oleic mutants (F435-2–1 and F435-2–2) were discovered with high oleic acid levels of 80% and low linoleic acid levels of 2%. This composition was similar to the oleic acid percentages found in olive oil (Norden et al., 1987). The oleic to linoleic (O/L) ratio in most commercial peanuts varies from 1.0–2.5 (Lopez et al., 2000); whereas, F435 has an O/L ratio near 35 (Norden et al., 1987). This high oleate phenotype originated from two recessive mutations in ahFAD2A and ahFAD2B (Moore and Knauft, 1989; Jung et al., 2000a), which seem to have no affect on other agronomic characteristics (Ray et al., 1993). These non-allelic genes are homoeologues originating from the diploid progenitors of A. hypogaea, encode microsomal oleoyl-PC desaturase also known as Δ12 fatty acid desaturase (Jung et al., 2000b). Normally, both of these homoeologous genes encode active desaturases (Bruner et al., 2001) that catalyze the first step in the biosynthesis of polyunsaturated fatty acids by converting oleic acid to linoleic acid by adding a second double bond in the hydrocarbon chain, which generates a polyunsaturated fatty acid from a monounsaturated fatty acid (Ray et al., 1993; Schwartzbeck et al., 2001). Loss of function of enzyme activity in both ahFAD2A and ahFAD2B has been shown to be responsible for the high oleate phenotype (Ray et al., 1993; Jung et al., 2000a).
Sequence analysis of ahFAD2A and ahFAD2B has revealed that the open reading frames (ORF) are 99% identical, encoding 379 amino acids, and contain no introns in the ORF (Jung et al., 2000b; Lopez et al., 2000). Comparison of a normal O/L and high O/L line uncovered four single nucleotide polymorphisms (SNPs) in these two homoeologous genes. Two SNPs were synonymous; whereas, the remaining SNPs were characterized by an insertion (442insA) generating a shift in the reading frame in ahFAD2B and a missense mutation (G448A or D150N) in ahFAD2A (Lopez et al., 2000; Yu et al., 2008). The presence or absence of the G448A (or D150N) point mutation in ahFAD2A was determined to be the key factor controlling the F2 segregation ratios of either 3∶1 or 15∶1, respectively, in crosses between normal and F435 derived high oleate peanuts (Jung et al., 2000a; Chu et al., 2007).
Growers have been demanding elite high oleic cultivars which can thwart disease and abiotic challenges that exist in different environments (Chu et al., 2007). Therefore, a concerted effort has been put forth to generate high oleic peanut lines with quality enhanced traits. In addition, more emphasis recently has been placed on growing high oleic peanuts by the peanut processing industry in the United States, Mexico, Australia, South Africa, and Brazil. Breeding multiple traits of interest such as disease resistance or other quality traits into high oleic cultivars is often time consuming due to the generation time, phenotyping for the traits of interest, and maintaining large populations that are needed to obtain desired trait stacking. Efficiency of peanut breeding programs can be greatly enhanced by developing DNA markers which are linked with traits of interest for marker assisted selection (MAS). Molecular markers can expedite this process by allowing the selection of progeny with the desired traits and removal of progeny that carry many undesirable traits at an early stage. The focus of this work was to employ a genotyping assay to rapidly detect ahFAD2B alleles (Barkley et al., 2010) in four segregation populations along with quantifying total fatty acid composition.
Materials and Methods
Parent Selection and Progeny Generation
Seeds of the parental lines were obtained from the USDA-ARS Plant Genetic Resources Conservation Unit in Griffin, GA. Two seeds per entry were germinated by planting them in a metal food serving tray containing a 1∶1 mixture of Metro-Mix 300 and perlite (Progress Growers Supply Inc. Ball Ground, GA). Plants were watered daily using an automatic watering system and daylight was extended by turning the greenhouse lights on from 6 pm to 11 pm. Greenhouse conditions were set to maintain temperatures between 21°C and 29.5°C. All crosses in this study were made during the summer of 2008.
Emasculation of flowers of the female parents started about 6 wk after planting. Emasculations were preformed in the evening between 6:30–8:00 pm. Wire ties were used to mark the emasculated flowers and aid in identification of the desired pegs. Non-emasculated flowers were removed every morning at 7am. Flowers from the male parents were selected the next morning and placed in vials with water. Pollinations were performed between 8:30–10:00 am. Pods were harvested 120 d after the last pollination except a cross involving PI 565455 Chico (an early maturing parent, Bailey and Hammons, 1975) which was harvested 90 d after the last pollination. All putative F1 seeds harvested were planted and grown in the greenhouse. Plants determined to be a product of self pollination based on the FAD2B genotyping assay were eliminated, while the hybrids were allowed to self and produce F2 seeds. The F2 seeds selected for further analysis were sliced with a razor blade (∼75–150 mg) distal from the embryo for DNA extraction and gas chromatography (GC). The remaining portion of the seed was treated with ethylene gas to germinate and the seedlings were established in the greenhouse for harvesting the next generation.
DNA Extraction and PCR Reactions
DNA samples were extracted by following the directions from an Omega-BioTek E.Z.N.A Plant DNA kit (Norcross, GA.). Leaf tissue or 75–150 mg of seed slices were used to extract DNA. Leaves or seed slices were placed in a 2 mL micro-centrifuge tube along with two 3 mm tungsten carbide beads (Qiagen Valencia, CA.) and 600 µl of P1 buffer from the Omega-BioTek kit. Tissue was pulverized by a Retsch Mixer Mill 301 (Leeds, UK) at 30 Hz for three minutes. Extracts were quantified on a DyNA Quant 200 fluorometer from Hoefer Pharmacia Biotech (San Francisco, CA). In addition, all samples were loaded on a 1% agarose gel (stained with ethidium bromide) along with a Low DNA Mass™ Ladder from Invitrogen (Carlsbad, CA) to evaluate quantity and determine the quality of each extraction. All samples were subsequently diluted to 10 ng/µl for Real-Time PCR.
The development of the genotyping assay, the PCR master mix, and cycling conditions were all as described previously (Barkley et al., 2010). All PCR reactions were performed in an ABI StepOne™ Real-time PCR machine using MicroAmp® fast optical 48-well plate and adhesive film seals (Applied Biosystems, Foster City, CA.). Each PCR run included non-template controls to ensure that reagents were free of contaminants. StepOne version 2.0 (Applied Biosystems, Foster City, CA.) was utilized to analyze and score genotypes among parents and progeny using the default parameters.
Gas Chromatography
Fatty acid composition was determined by gas chromatography on an Agilent 7890A (Agilent Technologies, Santa Clara, CA) gas chromatograph with a flame ionization detector (FID). Oil from a small amount (∼100 mg) of ground peanut seed was extracted in 5 mL of heptane and transesterified to fatty acid methyl esters (FAMEs) with 500 µl of 0.5 N sodium methoxide. Peak separation was performed on a DB-225 capillary column (15 m × 0.25 mm i.d. with a 0.25 µm film) from Agilent Technologies. One microliter of prepared sample was injected at a 60∶1 split ratio into the column maintained isothermally at 208°C. The inlet and detector were set at 280°C and 300°C, respectively. The carrier gas was helium set at a flow rate of 1 mL/min (38 cm/sec). Peaks were identified by comparison to a FAME standard mix RM-3 (Sigma-Aldrich, St Louis, MO). A total of eight fatty acids (palmitic C16∶0, stearic C18∶0, oleic C18∶1, linoleic C18∶2, arachidic C20∶0, gadoleic C20∶1, behenic C22∶0, and lignoceric acid C24∶0) were identified in each sample.
Data Analysis
GraphPad Prism version 3.0 was employed to statistically analyze the data and construct graphs. Correlations were determined by employing the Pearson correlation and calculating a two-tailed P value with 95% confidence intervals. One way ANOVA was utilized to test for significant differences among the mean values of oleic acid (C18∶1) for each genotypic class. Chi-square analysis was utilized to test the segregation patterns of digenic or monogenic inheritance for the oleic acid trait.
Results and Discussion
In order to evaluate the inheritance of ahFAD2B alleles and characterize fatty acid composition for each genotype, crosses were deliberately prepared between high oleic and normal oleic peanuts that were either early maturing such as PI 565455, Chico, (Bailey and Hammons, 1975) or previously displayed some disease resistance. The crosses chosen were cross number 17 [PI 652938 Florida-07 (Gorbet and Tillman, 2009)/PI 280688 A. hypogaea subsp. hypogaea var. hirsuta], cross number 19 [PI 653717 York/PI 502096 A. hypogaea subsp. fastigiata var. peruviana], cross number 25 [PI 651853 Tifguard (Holbrook et al., 2008)/PI 653717 York], and cross number 28 [PI 565455 Chico (Bailey and Hammons, 1975)/PI 653717 York]. [These selected genotypes have either a high oleic (ol1ol1ol2ol2) or normal oleic (ol1ol1Ol2Ol2 or Ol1Ol1Ol2Ol2) genotype which will produce a 3∶1 or 15∶1 segregation ratio of normal and high oleic F2 progeny, respectively. The first three crosses were selected because they either represent diverse germplasm, have disease resistance to TSWV, leaf spot, or nematodes. Cross 28 was selected because a previous study demonstrated that Spanish cultivars when crossed with a high oleic line did not always conform to either a digenic or monogenic inheritance pattern (Lopez et al., 2001).
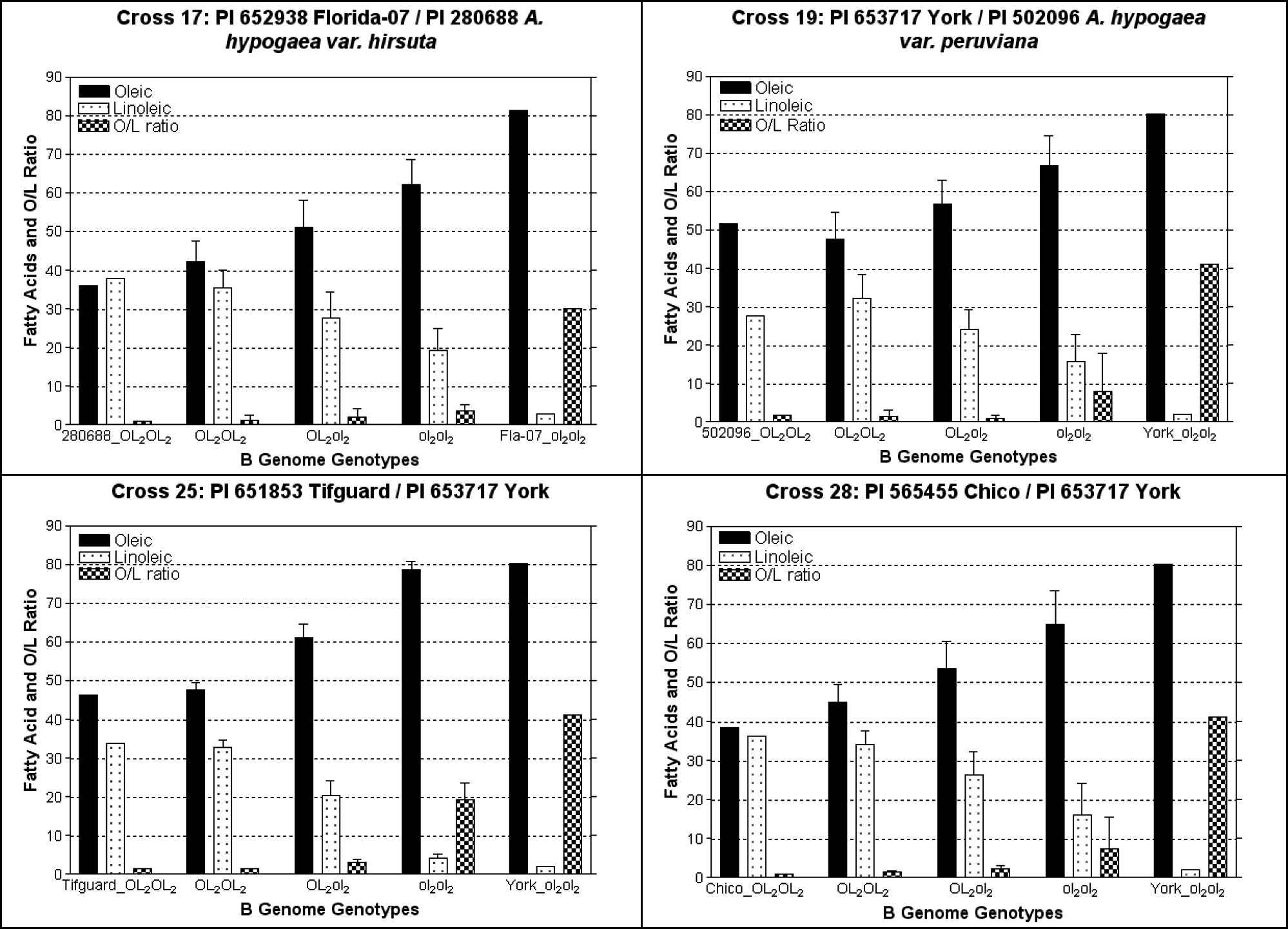
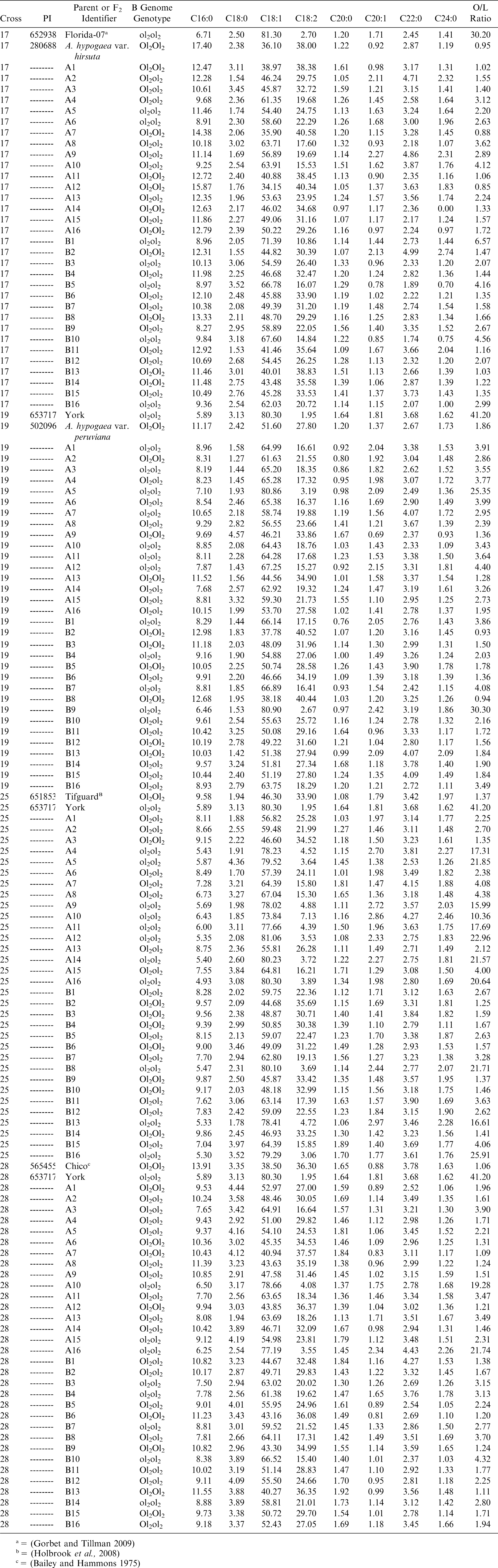
The oleic to linoleic ratio (O/L) of the parents was ascertained by gas chromatography (GC) and ranged from 0.95 to 41.2 and the genotype for ahFAD2B was determined by employing a Real-Time PCR genotyping assay to be either OL2OL2 or ol2ol2 (Table 1). A total of 84 seeds were obtained from the four crosses made. Leaves were harvested from all of the putative F1 progeny and subsequently genotyped for the 442insA mutation in ahFAD2B. The genotyping assay revealed that 75 plants (89.3%) were real F1 hybrids since they carried the ahFAD2B allele from the male parent; however, the remaining nine plants were products of self fertilization. The number of hybrids revealed here was slightly higher than a previous study which used microsatellite markers to distinguish hybrid from self fertilized progeny (Gomez et al., 2008). A total of 32 F2 seeds were randomly selected from each of these four crosses for ahFAD2B genotyping and fatty acid composition was collected (Table 1 and Fig. 1). An analysis of variance (ANOVA) revealed that the mean oleic acid produced from each genotypic class (Ol2Ol2, Ol2ol2, and ol2ol2) were significantly different at P < 0.0001. Overall, oleic acid (C18∶1) in these four F2 populations ranged from 34.15 to 81.06%; whereas, linoleic acid (C18∶2) ranged from 2.67 to 40.58%. The O/L ratio varied from 0.85 to 30.30. Although eight different fatty acids were detected, oleic (18∶1) and linoleic (18∶2) comprised the majority of the fatty acids found in the oil. This phenomenon has been previously documented in peanuts (Norden et al., 1987; Moore and Knauft, 1989).
Mean values of oleic, linoleic, and O/L ratio of the F2 progeny in selected crosses. (The parents of each cross are added on the far ends of the graph as a reference of normal and high oleic peanuts). The y-axis shows percentage of two fatty acids or total O/L ratio measured by GC and the x-axis shows the genotypes of F2 progeny and the parents. The F2 progeny are divided into the three detected ahFAD2B genotypes Ol2Ol2, Ol2ol2, and ol2ol2. ANOVA analysis demonstrated that the mean oleic values for each genotypic class were significantly different at P < 0.0001.
Phenotyping the fatty acid composition of the F2 populations revealed notable differences in these four crosses. First of all, cross 25 [PI 651853 Tifguard (Holbrook et al., 2008)/PI 653717 York], had a large portion of high oleic progeny produced from a relatively small sample set compared to the other three remaining crosses. For example, the majority (∼96%) of the F2 progeny produced from cross 17, 19, and 28 had O/L ratios less than 10; whereas, cross 25 had 34.4% of the F2 progeny with O/L ratios greater than 10 (Table 1, Fig. 1). The F2 individuals evaluated from cross 25 that were homozygous (ol2ol2) for the 442insA mutation in ahFAD2B all had oleic acid values ranging from 73.84 to 81.06% and O/L ratios ranging from 10.36 to 22.96. Progenies of the three remaining crosses that were homozygous (ol2ol2) for 442insA mutation in ahFAD2B had oleic acid values ranging from 54.4 to 80.90% and O/L ratios of 2.03 to 30.30. Further, each ahFAD2B genotype identified from cross 25 had a range of phenotypes produced for oleic (Ol2Ol2, 44.68 to 50.85; Ol2ol2, 55.81 to 67.04; and ol2ol2, 73.84 to 81.06) and linoleic acid (Ol2Ol2, 30.38 to 35.69; Ol2ol2, 15.30 to 26.28; and ol2ol2, 3.53 to 7.13) which were non-overlapping from one genotypic class to another; however, a similar trend was not observed in cross 17, 19, and 28 (Table 1 and Fig. 1). Consequently, crosses (17, 19, and 28) are still segregating for ahFAD2A so the F2 progeny with normal or mid-oleic acid phenotypes are expected to have at least one copy (or more) of the wild type allele in ahFAD2A, which causes the progeny to have lower oleic values and low O/L ratios.
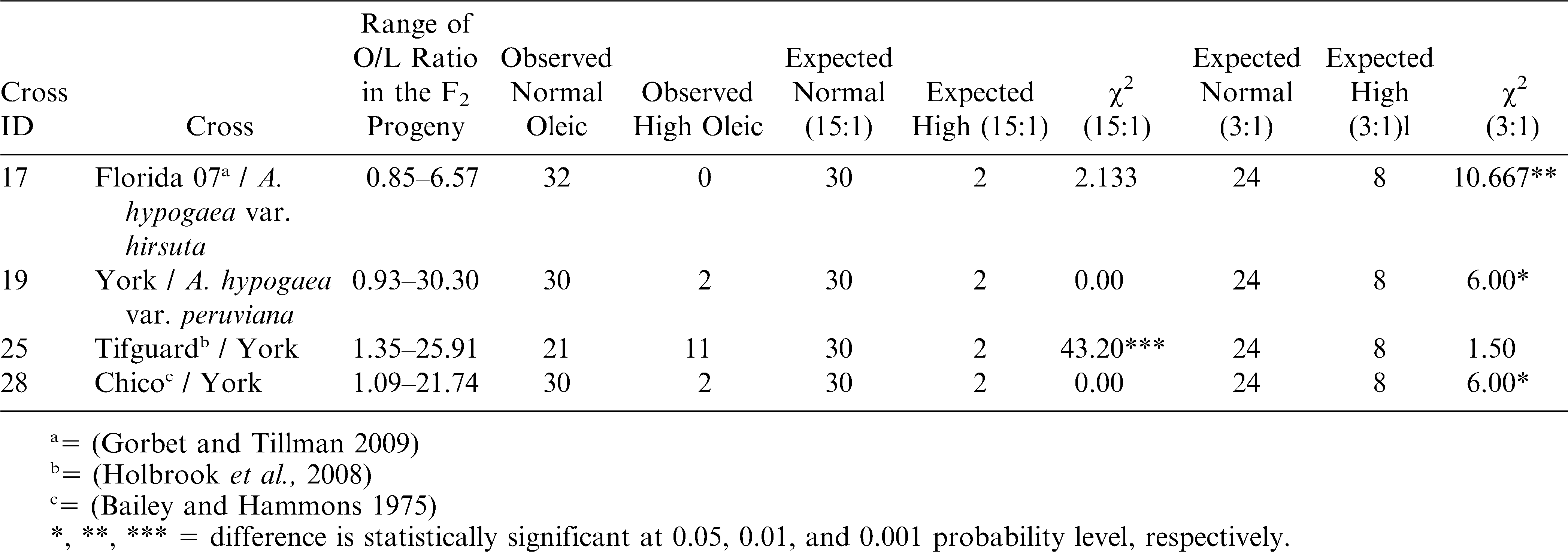
Previous reports have suggested that segregation ratios were generally either consistent with a monogenic 3∶1 or digenic 15∶1 inheritance for the oleic acid phenotype (Moore and Knauft, 1989; Lopez et al., 2001). The discrepancy in the segregation ratios for this trait was demonstrated to be controlled by the ahFAD2A genotype in the normal parent (Jung et al., 2000a; Chu et al., 2007). Hence, it was determined whether the segregation in these four F2 populations fit a monogenic or digenic inheritance pattern. Three of four crosses (17, 19, and 28) were consistent with a 15∶1 segregation ratio, but cross 25 conformed to a 3∶1 ratio (Table 2). None of these crosses evaluated had segregation ratios that did not fit any of the expected ratios as observed in a previous study (Lopez et al., 2001).
Observed and expected segregation ratios of high and low oleic acid (based on gas chromatography) in F2 peanut populations along with their χ2 values. All the χ2 values were compared for one degree of freedom. High oleic were classified as any progeny that had an O/L ratio > 10 and an oleic value > 70; whereas, normal oleic were classified as having an O/L ratio < 10 and oleic value < 70.
Several mid-oleic peanuts were observed in these segregating populations from the four crosses (Table 1). Genotypes observed for mid-oleic progeny were either heterozygous Ol2ol2 or homozygous recessive ol2ol2; although, the majority of the mid-oleic peanuts in the segregating populations had an ol2ol2 genotype. The F2 progeny, however, from cross 25 that were classified as mid-oleic, all had an Ol2ol2 genotype and putatively ol1ol1 in ahFAD2A. Though, in crosses 17, 19, and 28 mid oleic progeny had either an Ol2ol2 or an ol2ol2 genotype. This implies that mid oleic peanuts only have one copy of the wild type allele in either the A or B genome and three copies of the mutant allele to produce a mid-oleic phenotype. The genotypes (Ol1ol1ol2ol2 or ol1ol1Ol2ol2) would never breed true since segregation will occur in the next generation. However, it is also possible that mid-oleic cultivars have a genotype that is fixed for ahFAD2, but environmental effects or epistasis may result in an increase in C18∶1 to produce peanuts with 65 to 70% oleic acid. Prior studies have shown that oleate values can vary dependent on the genotype, seed maturity, and environmental interactions such as time of year planted, seasonal variation, insect damage, temperature, and soil conditions (Norden et al., 1987; Andersen and Gorbet, 2002). Further testing needs to be performed on mid-oleic peanuts to evaluate their genotype or determine if there are other genetic factors that control this phenotype.
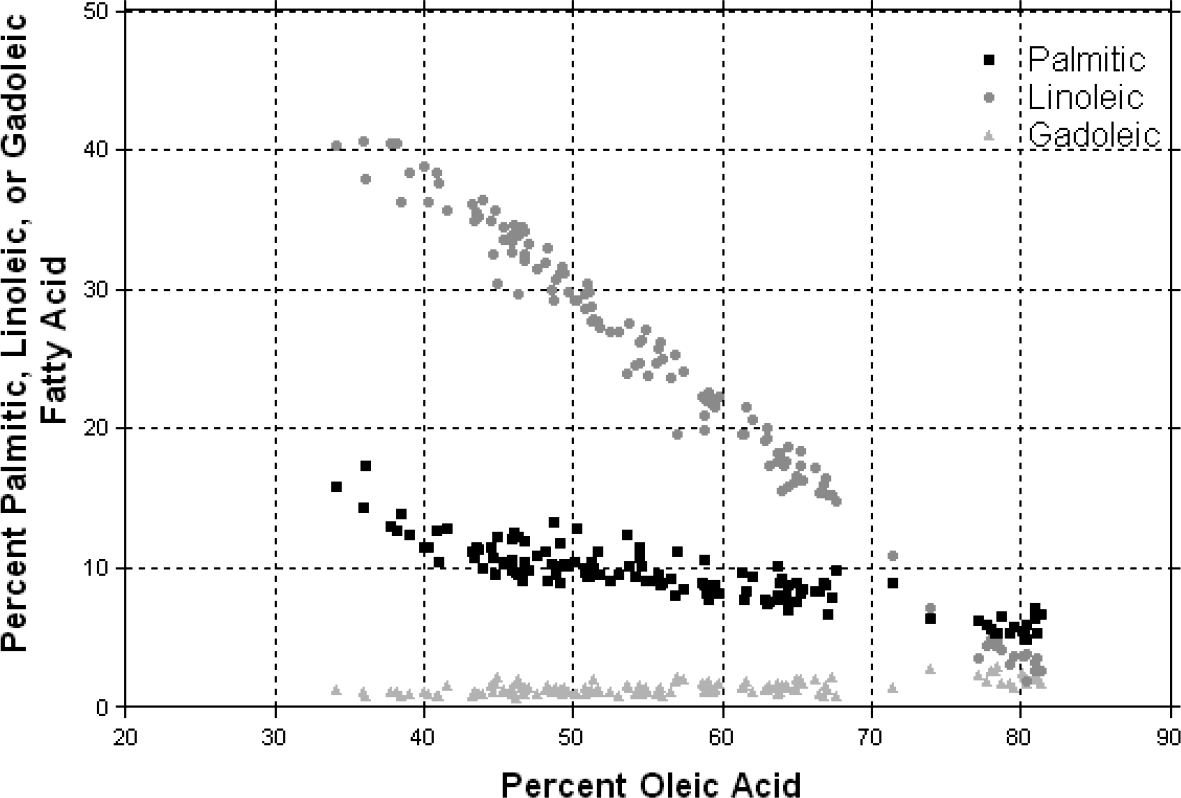
Significant correlations between oleic acid concentration and other fatty acids commonly detected in peanut have been previously reported (Isleib et al., 1996; Andersen and Gorbet, 2002; Isleib et al., 2006). Therefore, to determine whether different fatty acids vary together or independently, correlation coefficients were calculated utilizing the fatty acid composition measured in the segregating populations (Fig. 2). Oleic acid had a strong negative correlation with linoleic (r = − 0.9932; P < 0.0001) and palmitic acid (r = − 0.8796; P < 0.0001). On the other hand, a positive correlation (r = 0.6105; P < 0.001) was revealed between oleic and gadoleic acid (C20∶1). A weak, but, significant positive correlation (r = 0.2342; P < 0.001) was also found between oleic and lignoceric acid (C24∶0) [data not shown]. This suggests that the ol genotype influences the levels of oleic, linoleic, palmitic, and gadoleic fatty acids, and suggests that genetic modifiers may be involved for determining fatty acid composition. These data are comparable to correlations determined in previous studies (Isleib et al., 1996; Andersen and Gorbet, 2002; Isleib et al., 2006); however, weak correlations were detected previously between oleic and behenic acid (Andersen and Gorbet, 2002).
Development of cultivars with quality enhanced traits is often difficult and laborious, especially when plants must fully mature and produce seeds before assessing the presence or absence of the trait or traits of interest. The capability now exists to rapidly screen for the key mutations in ahFAD2 [CAPS markers previously developed for ahFAD2 (Chu et al., 2007; Chu et al., 2009) and 442insA in ahFAD2B via Real-Time PCR, (Barkley et al., 2010)], both of which are required for the high oleic phenotype in peanuts. These molecular assays can facilitate peanut breeding programs aiming to develop high oleate lines, by providing an early detection method of eliminating unwanted genotypes. Moreover, self fertilizalization can be rapidly identified and discarded from the population, which would save valuable time and space compared to growing out all the individual progenies. In time, more molecular markers will become available for selecting disease resistance and other novel traits; therefore, marker assisted selection (MAS) gene pyramiding may facilitate pyramiding multiple genes of interest in a single genotype (Kumar and Nayak, 2010), which will allow improved peanut breeding lines to be released in a shorter time period than conventional breeding.
Acknowledgements
The authors would like to thank Dr. Zhenbang Chen for comments to improve the manuscript. Also, thanks to Mr. Brandon Tonnis and Mr. Dave Pinnow for gas chromatography and DNA extraction assistance, respectively.
Literature Cited
Andersen P. C. and Gorbet D. W. 2002 Influence of year and planting date on fatty acid chemistry of high oleic acid and normal peanut genotypes. Jour. Ag. Food Chem 50 : 1298 – 1305 .
Bailey W. K. and Hammons R. O. 1975 Registration of Chico peanut germplasm (Reg. No. GP 2). Crop Sci 15 : 105 .
Barkley N. A. , Chenault Chamberlin K. D. , Wang M. L. , and Pittman R. N. 2010 Development of a real-time PCR genotyping assay to identify high oleic acid peanuts (Arachis hypogaea L.). Mol. Breed DOI 10.1007/s11032-009-9338-z.
Branch W. D. 1996 Registration of ‘Georgia Green’ Peanut. Crop Sci 36 : 806 .
Branch W. D. 2003 Registration of ‘GEORGIA-02C’ peanut. Crop Sci 43 : 1883 – 1884 .
Bruner A. C. , Jung S. , Abbott A. G. , and Powell G. L. 2001 The naturally occurring high oleate oil character in some peanut varieties results from reduced oleoyl-pc desaturase activity from mutation of aspartate 150 to asparagine. Crop Sci 41 : 522 – 526 .
Chu Y. , Ramos L. , Holbrook C. C. , and Ozias-Akins P. 2007 Frequency of a loss-of-function mutation in oleoyl-PC desaturase (ahFAD2A) in the mini-core of the US peanut germplasm collection. Crop Sci 47 : 2372 – 2378 .
Chu Y. , Holbrook C. C. , and Ozias-Akins P. 2009 Two alleles of ahFAD2B control the high oleic acid trait in cultivated peanut. Crop Sci 49 : 2029 – 2036 .
Dean L. L. , Henrix K. W. , Holbrook C. C. , and Sanders T. H. 2009 Content of some nutrients in the core of the core of the peanut germplasm collection. Peanut Sci 36 : 104 – 120 .
Gomez S. M. , Denwar N. N. , Ramasubramanian T. , Simpson C. E. , Burow G. , Burke J. J. , Puppala N. , and Burow M. D. 2008 Identifcation of peanut hybrids using microsatellite markers and horizontal polyacrylamide gel electrophoresis. Peanut Sci 35 : 123 – 129 .
Gorbet D. W. and Knauft D. A. 1997 Registration of ‘SunOleic 95R’ peanut. Crop Sci 37 : 1392 .
Gorbet D. W. and Tillman B. L. 2009 Registration of ‘Florida-07’ peanut. Jour. Plant Reg 3 : 14 – 18 .
Holbrook C. C. , Timper P. , Culbreath A. K. , and Kvien C. K. 2008 Registration of ‘Tifguard’ peanut. Jour. Plant Reg 2 : 92 – 94 .
Isleib T. G. , Wilson R. F. , and Novitzky W. P. 2006 Partial dominance, pleiotropism, and epistasis in the inheritance of the high oleate trait in peanut. Crop Sci 46 : 1331 – 1335 .
Isleib T. G. , Young C. T. , and Knauft D. A. 1996 Fatty acid genotypes of five Virginia-type peanut cultivars. Crop Sci 36 : 556 – 558 .
Jung S. , Powell G. , Moore K. , and Abbott A. 2000a The high oleate trait in the cultivated peanut [Arachis hypogaea L]. II. Molecular basis and genetics of the trait. Mol. Gen. Genet 263 : 806 – 811 .
Jung S. , Swift D. , Sengoku E. , Patel M. , Teule F. , Powell G. , Moore K. , and Abbott A. 2000b The high oleate trait in the cultivated peanut [Arachis hypogaea L.]. I. Isolation and characterization of two genes encoding microsomal oleoyl-PC desaturases. Mol. Gen. Genet 263 : 796 – 805 .
Kumar R. and Nayak S. 2010 Gene pyramiding-a broad spectrum technique for developing durable stress resistance in crops. Biotech. Mol. Bio. Review 5 : 51 – 60 .
Lopez Y. , Nadaf H. L. , Smith O. D. , Connell J. P. , Reddy A. S. , and Fritz A. K. 2000 Isolation and characterization of the Delta(12)-fatty acid desaturase in peanut (Arachis hypogaea L.) and search for polymorphisms for the high oleate trait in Spanish market-type lines. Theor. Appl. Genet 101 : 1131 – 1138 .
Lopez Y. , Smith O. D. , Senseman S. A. , and Rooney W. L. 2001 Genetic factors influencing high oleic acid content in Spanish market-type peanut cultivars. Crop Sci 41 : 51 – 56 .
Moore K. M. and Knauft D. A. 1989 The inheritance of high oleic acid in peanut. Jour. Hered 80 : 252 – 253 .
Norden A. J. , Gorbet D. W. , Knauft D. A. , and Young C. T. 1987 Variability in oil quality among peanut genotypes in the Florida breeding program. Peanut Sci 14 : 7 – 11 .
O'Byrne D. J. , Knauft D. A. , and Shireman R. B. 1997 Low fat-monounsaturated rich diets containing high-oleic peanuts improve serum lipoprotein profiles. Lipids 32 : 687 – 695 .
Parthasarathy S. , Khoo J. C. , Miller E. , Barnett J. , Witztum J. L. , and Steinberg D. 1990 Low density lipoprotein rich in oleic acid is protected against oxidative modification: Implications for dietary prevention of atherosclerosis. Proc. Natl. Acad. Sci. USA 87 : 3894 – 3898 .
Pelkman C. L. , Fishell V. K. , Maddox D. H. , Pearson T. A. , Mauger D. T. , and Kris-Etherton P. M. 2004 Effects of moderate-fat (from monounsaturated fat) and low-fat weight-loss diets on the serum lipid profile in overweight and obese men and women. Amer. Jour. Clin. Nutr 79 : 204 – 212 .
Ray T. K. , Holly S. P. , Knauft D. A. , Abbott A. G. , and Powell G. L. 1993 The primary defect in developing seed from the high oleate variety of peanut (Arachis hypogaea L.) is the absence of delta 12-desaturase activity. Plant Sci 91 : 15 – 21 .
Schwartzbeck J. L. , Jung S. , Abbott A. G. , Mosley E. , Lewis S. , Pries G. L. , and Powell G. L. 2001 Endoplasmic oleoyl-PC desaturase references the second double bond. Phytochemistry 57 : 643 – 52 .
Teres S. , Barcelo-Coblijn G. , Benet M. , Alvarez R. , Bressani R. , Halver J. E. , and Escriba P. V. 2008 Oleic acid content is responsible for the reduction in blood pressure induced by olive oil. Proc. Natl. Acad. Sci. USA 105 : 13811 – 6 .
Vassiliou E. K. , Gonzalez A. , Garcia C. , Tadros J. H. , Chakraborty G. , and Toney J. H. 2009 Oleic acid and peanut oil high in oleic acid reverse the inhibitory effect of insulin production of the inflammatory cytokine TNF-alpha both in vitro and in vivo systems. Lipids Health Dis 8 : 25 .
Yu S. , Pan L. , Yang Q. , Min P. , Ren Z. , and Zhang H. 2008 Comparison of the Delta(12) fatty acid desaturase gene between high-oleic and normal-oleic peanut genotypes. Jour. Genet. Genomics 35 : 679 – 685 .
Zhang D. , Pirtle I. L. , Park S. J. , Nampaisansuk M. , Neogi P. , Wanjie S. W. , Pirtle R. M. , and Chapman K. D. 2009 Identification and expression of a new delta-12 fatty acid desaturase (FAD2-4) gene in upland cotton and its functional expression in yeast and Arabidopsis thaliana plants. Plant Physiol. Biochem 47 : 462 – 471 .
Notes
- USDA-ARS, Plant Genetic Resources Conservation Unit, 1109 Experiment Street, Griffin, GA 30223 USA. [^]
- USDA-ARS, Wheat, Peanut, and Other Field Crops Research Unit, 1301 N. Western Stillwater, OK 74075 USA. [^] *Corresponding author, Phone: 770-412-4035, Fax: 770-229-3323, E-mail: Elle.Barkley@ars.usda.gov
Author Affiliations