Introduction
Peanut (Arachis hypogaea L.) is one of the most susceptible host crops to Aspergillus flavus (Link) invasion and subsequent aflatoxin production before and after harvest. The extent of aflatoxin contamination varies with geographic location, agricultural and agronomic practices, storage and processing period and conditions (Stoloff, 1985). In China, the severity of aflatoxin contamination gradually decreases as latitude increases. Contamination is more serious in southern peanut production areas than in northern areas (Xiao, 1989). The Chinese government pays great attention to the aflatoxin contamination problem and the research projects in this field receive high priority. Adopting new cultural, curing and storage practices can minimize aflatoxin contamination. However, these practices may not be suited to small-scale farming in developing countries, especially in tropical areas. Chemical control and removal of toxins have not yet been completely successful. An effective solution to the problem may be the use of peanut varieties that are resistant to infection by the aflatoxin-producing fungi, or varieties that suppress aflatoxin production if colonized by the fungus (Anderson et al., 1995). Advances in genomic tools and information through the international Peanut Genomics Initiative should accelerate the development of agronomically acceptable peanut cultivars with reduced aflatoxin contamination.
In this review, recent advances are highlighted concerning host resistance against aflatoxin contamination and genomics research activities conducted at the Crops Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China.
Host Resistance to Aflatoxin Contamination
Identification of resistant germplasm and development of resistant cultivars
Through screening of over 2,000 germplasm accessions of peanuts, 20 genotypes were identified as highly resistant to A. flavus invasion in seeds (Zhou et al., 2002). These resistant genotypes include local landraces and some genotypes introduced from International Crops Research Institute for the Semi-Arid Tropics (ICRISAT). PI 337494F, J-11, Zhanqiu 48, UF71513 and Meixianhonhyi have also been used in the aflatoxin resistance breeding program. Two improved cultivars Yueyou 9 and Yueyou 20 with high resistance to A. flavus invasion and acceptable agronomic traits were released in 2004 by the National Peanut Varieties Approval Committee and Guangdong Crops Varieties Approval, respectively. Some promising advanced breeding lines with resistance to preharvest aflatoxin contamination (50 to 66% reduction in aflatoxin contamination compared to susceptible accessions in the fields) also have been developed. Zhou et al. (1999, 2002) studied the inheritance of resistance to seed infection by A. flavus and found the resistant characters were controlled by a pair of major genes with additive value 0.38 and a pair of minor genes with additive value 0.12. The additive gene actions were important for resistant, and the percent of recombination between parent genotypes was estimated at 43.22%. Heritability was estimated at 58%.
Identification of resistance traits
A better understanding of mechanisms of resistance to fungal infection and aflatoxin production should accelerate the development of resistant cultivars. Mechanisms of peanut resistance to A. flavus invasion and aflatoxin production have been studied at the Guangdong Academy since 1999 and several significant factors have been observed.
Structure of seed coat
Liang et al. (2003b) studied the role of wax and cutin layers in peanut seed coats in resistance to invasion and colonization by A. flavus. Results showed that wax contents of resistant genotypes were significantly higher than susceptible cultivars. The resistant kernels had a thicker and coarser waxy deposit on seed coat surfaces than susceptible genotypes as observed by scanning electron microscope. Removal of wax with chloroform or removing of cutin with KOH and cutinase can increase the susceptibility of peanut seeds. The bioassays of wax in vitro showed that there were no significant test-by-treatment interactions. These results indicated that the wax and cutin layers of peanut seed coat might only be a physical barrier to A. flavus invasion and colonization.
Active oxygen and membrane lipid peroxidation
Liang et al (2002) observed differences in the changes of active oxygen species including superoxide anion (O2-), hydrogen peroxide (H2O2), and hydroxyl radial (OH-), lipoxygenase (LOX) activity and membrane lipid peroxidation levels between resistant and susceptible seeds after inoculation with A. flavus. In resistant genotypes, the levels of malondialdehyde and the degree of membrane lipid peroxidation were significantly increased (7-8×) at 2-3 d after inoculation. Moreover, the generation rates of O2-, H2O2, and LOX were also increased markedly at an early stage after inoculation. O2- and H2O2 were increased rapidly and quickly reached a maximum level. However, no significant increases in the activities of superoxide dismutase (SOD) or catalase (CAT) were observed, which implies that there was not enough SOD and CAT to scavenge active oxygen. The accumulation of active oxygen and the increased activity of LOX can cause changes in membrane lipid peroxidation, cell wall strength, synthesis of phytoalexin and hypersensitive cell death (Dixion and Harrison, 1994; Baker and Orlandi, 1995). In contrast, the MDA(Malondialdehyde)level was increased at 5-6 d, and the production rates of O2-, content of H2O2, and activity of LOX in susceptible genotypes was increased much later than in the resistant genotypes (Liang et al., 2001, 2002).
Phytoalexin accumulation
Phytoalexins are antibiotic secondary metabolites produced by plants in response to injury and invasion by some pathogens and appear to be involved in disease resistance (Keen, 1986; Sobolev et al., 2007). Resveratrol is one of the phytoalexin compounds found in peanut seeds (Sanders et al., 2000). Liang et al. (2006b) compared the synthesis capacity for resveratrol between the resistant and susceptible seeds after inoculation with A. flavus. The results showed that the accumulation of resveratrol in resistant genotypes was increased 3× at 3 d after inoculation, but susceptible genotypes did not reach the same levels until 4 d after inoculation.
Antifungal proteins
Protein profiles of 15 peanut genotypes revealed that the trypsin inhibitor and lipid transfer protein were present at relatively high concentration in resistant genotypes (Liang et al., 2003a, 2003c, 2004b). Both proteins exhibited strong bioactivity against the growth of A. flavus. In another investigation, the difference of total seed protein in resistant and susceptible peanut genotypes was investigated by proteomic approaches (Liang et al., 2004a). The major qualitative difference between resistant genotypes and susceptible genotypes is that resistant genotypes contained three unique proteins P1 (22.5 kD, pI 4.1), P2 (22.5 kD, pI 8.2) and P3 (23.8 kD, pI 5.9), while susceptible genotypes contained one unique protein P4 (23.5 kD, pI 7.0). Another protein P5 (22.5 kD, pI 7.3) also was found in concentration tenfold more in resistant vs. susceptible genotypes. The peptide sequences of spots P-1 and P-3 are identical to legumin A precursor from Vicia narbonensis L. The peptide sequences of spots P-2 and P-4 are the same and identical to glycinin from A. hypogaea. The polymorphic protein peptides distinguished by 2-D PAGE may be used as markers for identification of resistant peanut lines. In addition to constitutive seed antifungal proteins, there were significant differences of two pathogenesis-related proteins (chitinase and 𝛃-1-3-glucanase) between the resistant and susceptible genotypes after inoculation with A. flavus (Liang et al., 2003a, 2005). The activities of endo-chitinase and 𝛃-1-3-glucanase increased earlier in resistant than in susceptible genotypes after invasion by A. flavus, while more isoform bands of 𝛃-1-3-glucanase were observed in resistant than in susceptible genotypes. The purified chitinase can significantly inhibit spore germination and hypha growth of A. flavus in vitro, while thin-layer chromatography analysis of the hydrolytic product from 𝛃-1-3-glucanase and hypha of A. flavus revealed the presence of enzymatic hydrolytic oligomer products (Liang et al., 2005).
Recent Progress in Peanut Genomic Research
Genetic diversity in cultivated peanuts based on SSR markers
A genetic diversity analysis of 28 accessions of A. hypogaea, consisting of seven accessions of var. fastigiata, five accessions of var. hirsuta, eight accessions of var. hypogaea and eight accessions of var. vulgaris, was performed with 110 SSR primer pairs (Hong et al., 2008). Forty six primer pairs detected polymorphisms in these varieties with two to nine alleles per locus. The value of PIC (polymorphism information content) varied from 0.080 to 0.869. Clustering analysis indicated that classification of these varieties was in general agreement with traditional taxonomic classification based on DNA fingerprints. However, the varieties which are var. fastigiata according to traditional taxonomic classification couldn't be clustered into the same group on the basis of these molecular criteria. At the same time, a SSR marker (pPGSseq15C12) that could differentiate var. vulgaris from the other varieties was identified. The PCR polymorphic bands amplified by primer pPGSseq15C12 were further characterized via cloning and sequencing. The results showed that the polymorphism between var. vulgaris and the other varieties was due to the different repeat number of the ATT motif. These results suggest that SSR markers should be useful tools in future studies on genetic diversity at the molecular level.
Mining and characterization of peanut EST-SSR
SSR markers have potential use for genetic studies of the cultivated peanut. Unfortunately, SSR markers have not been fully developed in cultivated peanut and the development of traditional genomic SSRs from genomic DNA is costly and time-consuming. EST-SSR represents functional molecular markers as "putative function" for the majority of such markers that can be deduced by database searches and other in silico approaches. Furthermore, EST-SSR markers are expected to possess high interspecific transferability as they belong to relatively conserved genomic regions. With the recent increased numbers of ESTs deposited in the database in public Genbank, it is possible to identify and develop EST-SSR markers on a large scale in a timely and cost-effective manner. Due to the above advantages of EST-SSR markers and relatively easy accessibility of large EST resources, large numbers of EST-SSR markers are being identified and used for a number of applications in many plant species (Gupta et al., 2003, Guo et al., 2006). To date, however, little information on EST-SSR markers in peanut is available. In our investigation (Chen et al., 2006), a total of 24,238 peanut EST sequences available in public database were retrieved for development of EST-SSRs through data mining. After an extensive computational analysis, these ESTs resulted in 11,431 potentially unique ESTs with 1,434 contigs and 9,997 singletons, from which about 6.8% SSR-containing EST sequences, including 881 EST-SSRs equivalent to an average of one SSR in every 7.3 kb. Among these EST-SSRs with a motif length of 2-6 bp, approximate 63.9% are tri-nucleotide, followed by di- (32.7%), tetra- (1.7%), hexa- (1%), and penta-nucleotide (0.7%) motif types. Among them AG/TC, AAG/TTC, AAT/TTA, ACC/TGG, ACT/TGA and AT/AT ranked as the top 6 motif types, frequency of which considering sequences complementary was more than 6%. Only 290 primer pairs were designed from the 881 EST-SSRs and used to examine polymorphism among 22 accessions of A. hypogaea representing subspecies hypogaea and fastigiata. In addition, 15 accessions of wild Arachis species were analyzed. Within these primers, 86.6% produced amplification products, of which 10% and 88% resulted in polymorphism among the cultivated and wild species, respectively. Alleles varying from two to four were found among cultivated peanuts, whereas three to eight alleles were present among wild species. Sequences of the PCR polymorphic bands indicated that insertions or deletions occurred in SSR sites among alleles of wild species and cultivated peanuts. These results suggest that peanut EST-SSR markers can enhance the current resource of molecular markers in peanuts. Furthermore, EST-SSRs markers have considerable potential for genetic mapping, and could be used in quantitative trait loci (QTLs) mapping (Table 1) and marker assisted selection (MAS).
Transferability of soybean EST-SSR in cultivated peanut
As EST-SSR markers from transcribed regions of DNA are generally more conserved across species than genomic SSR markers from un-transcribed regions, EST-SSRs have been demonstrated to be highly transferable among species (Gupta et al., 2003), implying that they may be of significant value in comparative mapping and evolutionary analysis among taxa. Recently, relatively high transferability of soybean genomic-SSR has been observed in peanut (He et al., 2006). However, the transferability of EST-SSR derived from soybean is unknown. Our goal was to develop EST-SSR from soybean and to examine the transferability in peanut. A total of 2082 EST-SSRs were identified from 391,578 soybean EST sequences retrieved from the GenBank. The bi-nucleotide repeat motif was the most abundant type of SSRs (63.5%), followed by tri- (30.9%); hexa- (2.5%), tetra- (2%) and penta-nucleotide (1.1%) repeat motfs. From the 2082 SSR-containing EST sequences, 685 primer pairs were successfully designed. Among these ESTs, 228 (33.3%) were found in 5'-untranslated terminal region (5'-UTR), 300 (43.9%) in translated region, and 157 (22.8%) in 3'-UTR (Chen et al., 2006b).
The transferability of 685 soybean-derived EST-SSR primers was assessed among 22 cultivated genotypes representing four market types and 16 wild species including A- and B-genome species. Results of PCR analysis showed that only 78 of the 685 soybean EST-SSR could produce amplification products in both wild and cultivated peanut, and 596 could detect amplification products in Dongnong 594, a soybean line as control. This indicated that these soybean-derived EST-SSRs have a low rate of transferability (11.3%) across species in both wild and cultivated peanut. The SSR banding patterns produced by these primers were different between peanut and soybean. Among these transferable soybean EST-SSRs, 77 (98.7%) and 20 (25.3%) SSR primers detected polymorphism among wild and cultivated peanuts, respectively. The number of alleles detected by these primers varied from two to five and from two to seven in cultivated and wild peanut, respectively.
Genetic linkage map and QTL analysis
Molecular genetic maps of crop species can be used in a variety of ways in breeding and genomic research such as identification and mapping of genes and quantitative trait loci (QTLs) for morphological, physiological, and economic traits of crop species. However, a comprehensive genetic linkage map for cultivated peanut has not yet been developed due to extremely low levels of DNA polymorphism in cultivated peanut (Stalker and Mozingo, 2001). Recently, a total of 1043 SSR primer pairs including 638 genomic-SSRs and 406 EST-SSRs were tested in four parental lines, in which three independent recombinant inbred line (RIL) mapping populations (Yueyou 13 × Zhenzhuhei, Yueyou13 × Fu95-5 and Yueyou13 × J-11 ) were developed. Among them are 143 SSR markers that express polymorphisms between Yueyou13 and Zhenzhuhei with 145 loci, 123 SSRs between Yueyou13 and Fu95-5 with 125 loci, and only 59 SSRs between Yueyou13 and J-11. The polymorphic SSR markers were used to construct three independent maps with 138, 108 and 42 markers, respectively, based on 392 F5 individuals derived from these three RIL mapping populations. An integrated genetic linkage map was constructed by combing these three mapping populations using the computer program JoinMap® 3.0 (Hong et al., unpublished data). This integrated map consists of 179 SSR loci in 21 linkage groups and covered 973.4 cM, with an average of 5.8 cM of intermarker distance. In addition, one SSR marker PM93/630-600 was linked to peanut testa color (Hong et al., 2007). This SSR-based genetic linkage map contains 18 functional marker loci (EST-SSR loci) (Hong et al., 2006).
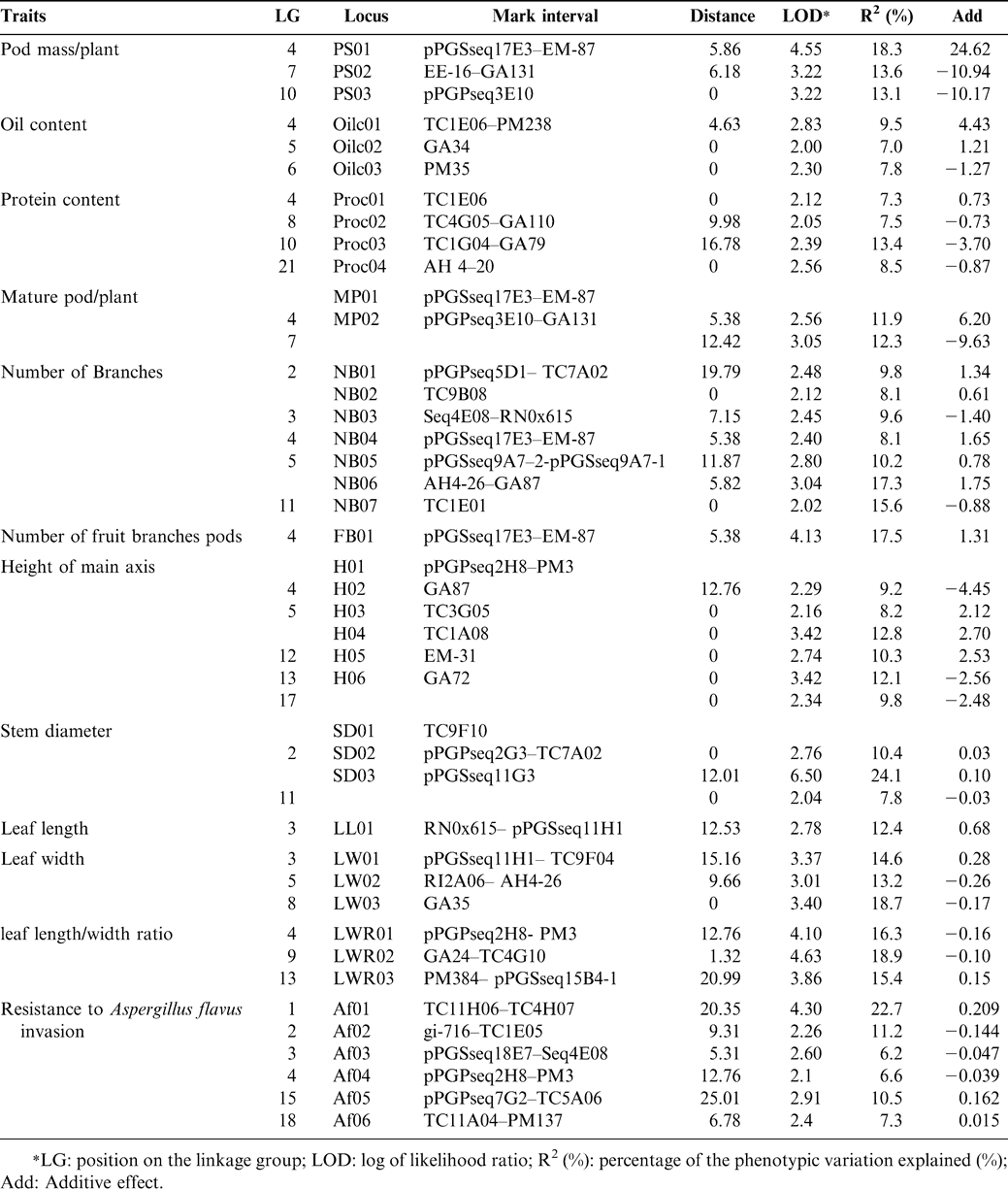
Based on the newly developed linkage maps, we have conducted several QTL linkage analyses for height of main axis, pod weight/plant, number of mature pods/plant, stem diameter, number of total branches, number of fruiting branches, leaf length, leaf width, leaf length/width ratio, protein content, oil content, and resistance to A. flavus infection. The position on a linkage group (LG), log of likehood ratio, marker interval, distance, percentage of the phenotypic variation explained, and additive effect of QTLs associated with 12 traits were determined (Table 1). A total of 42 QTLs have been detected that explained 7.0-24.1% of the total variation for different traits, and one to seven QTLs were identified for each of the traits. This information should be useful in future studies on peanut breeding and genetics.
Conclusions
Resistance mechanisms in peanut to aflatoxin contamination have been studied for several years. Resistance mechanisms and factors have been reported to contribute to suppression of A. flavus, but no effective efforts have been made to select for these traits. Information about mechanisms of resistance is not completely clear because of high G × E interactions. As limited progress has been made in the molecular analyses, future research will focus on providing a comprehensive understanding of the functions of peanut genes by using gene expression analysis tools. A better understanding and use of resistance mechanisms should result from advanced genomic tools and peanut specific bio-information.
Acknowledgements
This research was partially funded by a grant from National Natural Science foundation of China (No 30571179) and funds provided by National "863"project of China (No 2006AA0Z156, 2006AA10A115). The authors also express gratitude to Drs. Baozhu Guo and Corley Holbrook, USDA-ARS, Tifton, GA for helpful discussions.
Literature Cited
Anderson W. F., Holbrook C. C., Wilson D. M., and Matheron M. E. . 1995. Evaluation of preharvest aflatoxin contamination in some potentially resistant peanut genotypes. Peanut Sci 22: 29- 32.
Baker C. J. and Orlandi E. W. . 1995. Active oxygen in plant pathogenesis. Annu. Rev. Phytopathol 33: 299- 321.
Dixon R. A. and Harrison M. J. . 1994. Early events in the activation of plant defense responses. Ann. Rev. Phytopathol 32: 479- 501.
Guo W., Wang W., Zhou B., and Zhang T. . 2006. Cross-species transferability of G. arboreum-derived EST-SSRs in the diploid species of Gossypium. Theor. Appl. Genet 112: 1573- 1581.
Gupta P. K., Rustgi S., Sharma S., Singh R., Kumar N., and Balyan H. S. . 2003. Transferable EST-SSR marker for the study of polymorphism and genetic diversity in bread wheat. Mol. Gen. Genomic 270: 315- 323.
He G. H., Woulhard F. E., Marong I., and Guo B. Z. . 2006. Transferability of soybean SSR markers in peanut (Arachis hypogaea L.). Peanut Sci 33: 22- 28.
Hong Y. B., Chen X. P., Liang X. Q., and Liu H. Y. . 2008. Genetic diversity analysis in cultivated peanut (Arachis hypogaea L.) based on SSR polymorphism. Mol. Plant Breeding 6/ 1: 71- 78.
Hong Y. B., Lin K. Y., Zhou G. Y., Li S. X., Li Y., and Liang X. Q. . 2007. Genetic linkage analysis of SSR markers and the gene for dark purple testa color in peanut (Arachis hypogaea L.) Chinese. Jour. Oil Crop Sci 29: 35- 38.
Keen N. T. 1986. Phytoalexin and their involvement in plant disease resistance. Iowa State Jour. Res 60: 477- 499.
Liang X. Q., Guo B. Z., and Holbrook C. C. . 2004a. Identification of peanut seed-storage proteins associated with resistance against Aspergillus flavus infection and aflatoxin production (abstr.). Proc. Amer. Peanut Res. and Educ. Soc 36: 74- 75.
Liang X. Q., Guo B. Z., Holbrook C. C., and Lynch R. E. . 2003a. Resistance to Aspergillus flavus in peanut seeds is associated with constitutive trypsin inhibitor and inducible chitinase and 𝛃-1-3-glucanase (abstr.). Proc. Amer. Peanut Res. and Educ. Soc 35: 85.
Liang X. Q., Holbrook C. C., Lynch R. E., and Guo B. Z. . 2005. 𝛃-1,3-glucanase activity in peanut seed (Arachis hypogaea L.) is induced by inoculation with Aspergillus flavus and copurifies with a conglutin-like protein. Phytopathology 95: 506- 511.
Liang X. Q., Pan R. Z., and Zhou G. Y. . 2002. Involvement of active oxygen generation and lipid peroxidation in the susceptibility/resistance of peanut seed infected by Aspergillus flavus. Chinese Jour. Oil Crop Sci 24: 14- 17.
Liang X. Q., Zhou G. Y., and Pan R. Z. . 2001. Changes of some biochemical substances in peanut seeds under infection of Aspergillus flavus and their role in resistance to seed invasion. Chinese Jour. Oil Crop Sci 23: 26- 31.
Liang X. Q., Zhou G. Y., and Pan R. Z. . 2003b. Study on the relationship of wax and cutin layers in peanut seeds and resistance to invasion and aflatoxin production by Aspergillus flavus. Jour. Tropic. and Subtropical Bot 11: 11- 14.
Liang X. Q., Zhou G. Y., and Pan R. Z. . 2003c. Relationship of trypsin inhibitor in peanut seed and resistance to Aspergillus flavus invasion. Acta Agronomica Sinica 29: 23- 28.
Liang X. Q., Zhou G. Y., and Zou S. C. . 2006b. Differential induction of resveratrol in resistant peanut seeds infected by Aspergillus flavus. Chinese. Jour. Oil Crop Sci 28: 63- 66.
Sanders T. H., McMichael R. W., and Hendrix K. W. . 2000. Occurrence of resveratrol in edible peanut. Jour. Agric. Food Chem 48: 1243- 1246.
Sobolev V. S., Guo B. Z., Holbrook C. C., and Lynch R. E. . 2007. Interrelationship of phytoalexin production and disease resistance in selected peanut genotypes. Jour. Agric. Food Chem 55: 2195- 2200.
Stalker H. T. and Mozingo L. G. . 2001. Molecular markers of Arachis and marker-assisted selection. Peanut Sci 28: 117- 123.
Zhou G. Y. and Liang X. Q. . 1999. Studies on the ultramicroscopic structure of seed coats between resistant and susceptible to Aspergillus flavus invasion in peanut. Chinese Jour. Oil Crop Sci 21: 32- 35.
Zhou G. Y. and Liang X. Q. . 2002. Analysis of major-minor genes related to resistance to infection by Aspergillus flavus in peanut. Jour. of Peanut Sci 31: 11- 14.
Zhou G. Y., Liang X. Q., Li Y. C., Li X. C., and Li S. L. . 1999. Genetic analysis of traits of resistance to Aspergillus flavus in peanut. Jour. of Peanut Sci 4: 140- 143.
Zhou G. Y., Liang X. Q., Li Y. C., Li X. C., and Li S. L. . 2002. Evaluation and application of introduced peanut cultivars for resistance to Aspergillus flavus invasion. Jour. of Peanut Sci 32: 14- 17.
Notes
Author Affiliations
1 Crops Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, Guangdong 510640, China.
*Corresponding author's email: Liang804@yahoo.com